-
Using Clinical Data to Embed Patients
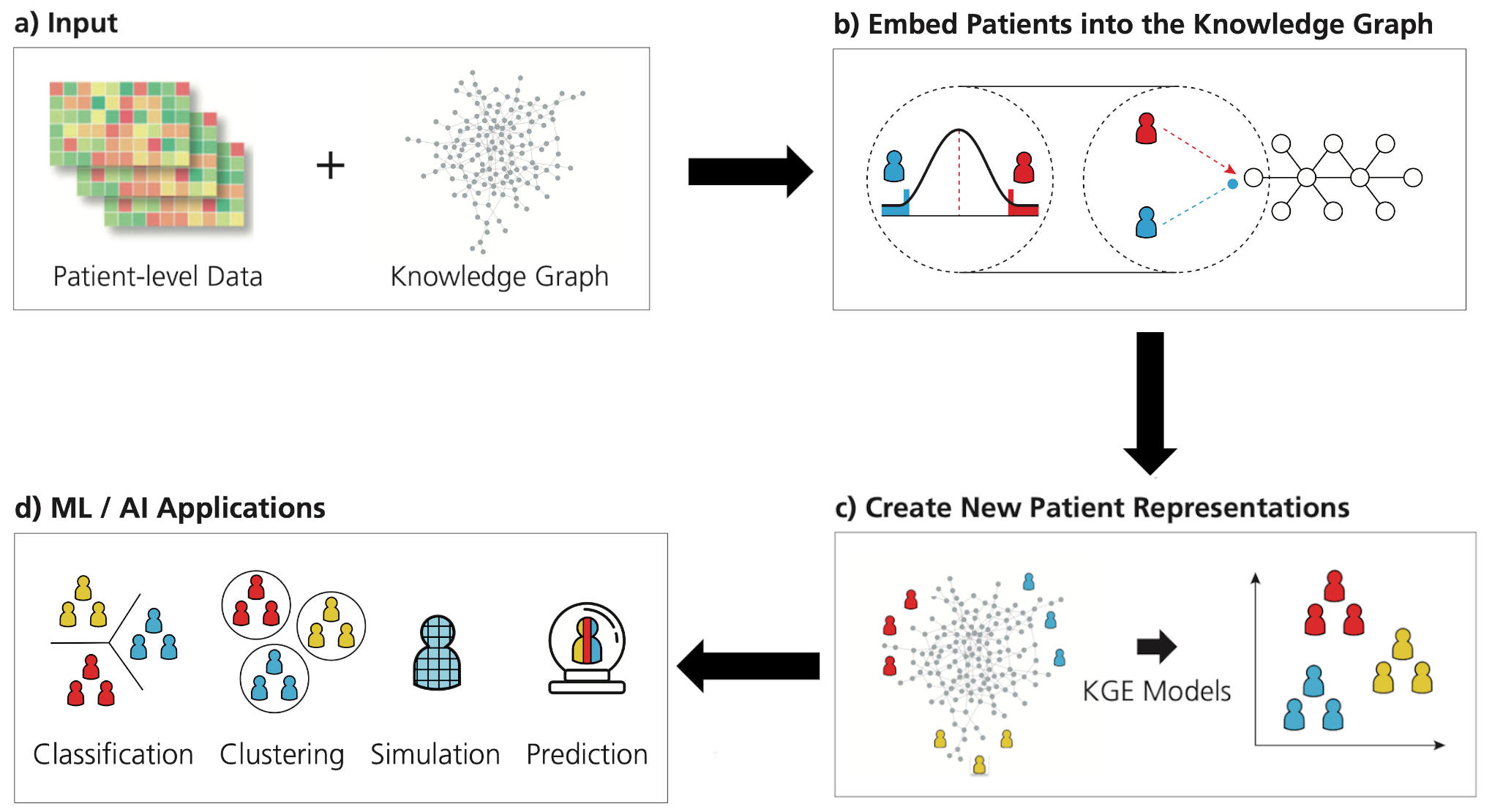
The expression of each gene is often measured in groups of patients with a given disease to compare to healthy patients. It is then calculated which genes are higher, lower, or similar to healthy patients. We’ve used these calculations to introduce patients into a biomedical knowledge graph containing genes so we could generate an embedding for each patient using PyKEEN. After, we showed these embeddings are useful for classifying new patients and other downstream ML tasks.
-
Benchmarking Study
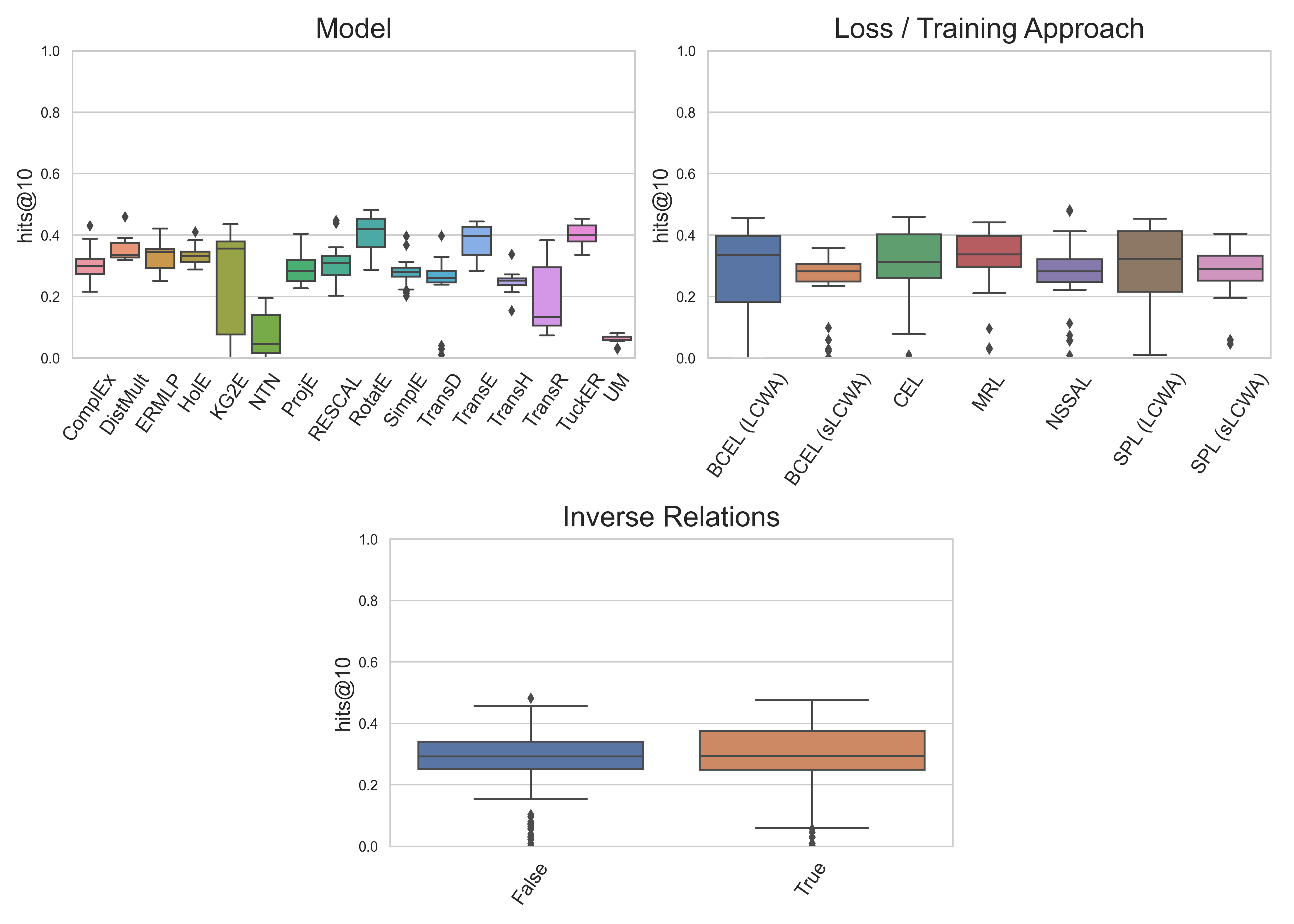
We’ve run an unprecedented large benchmarking study. This image describes the results on the FB15k237 dataset across several knowledge graph embedding models, loss functions, training approaches, and usages of explicit modeling of inverse triples. This is just one of several datasets analyzed in this study. In our manuscript, we also assess the reproducibility of old models’ best reported hyperparameters.
-
Metaresearch Recommendations
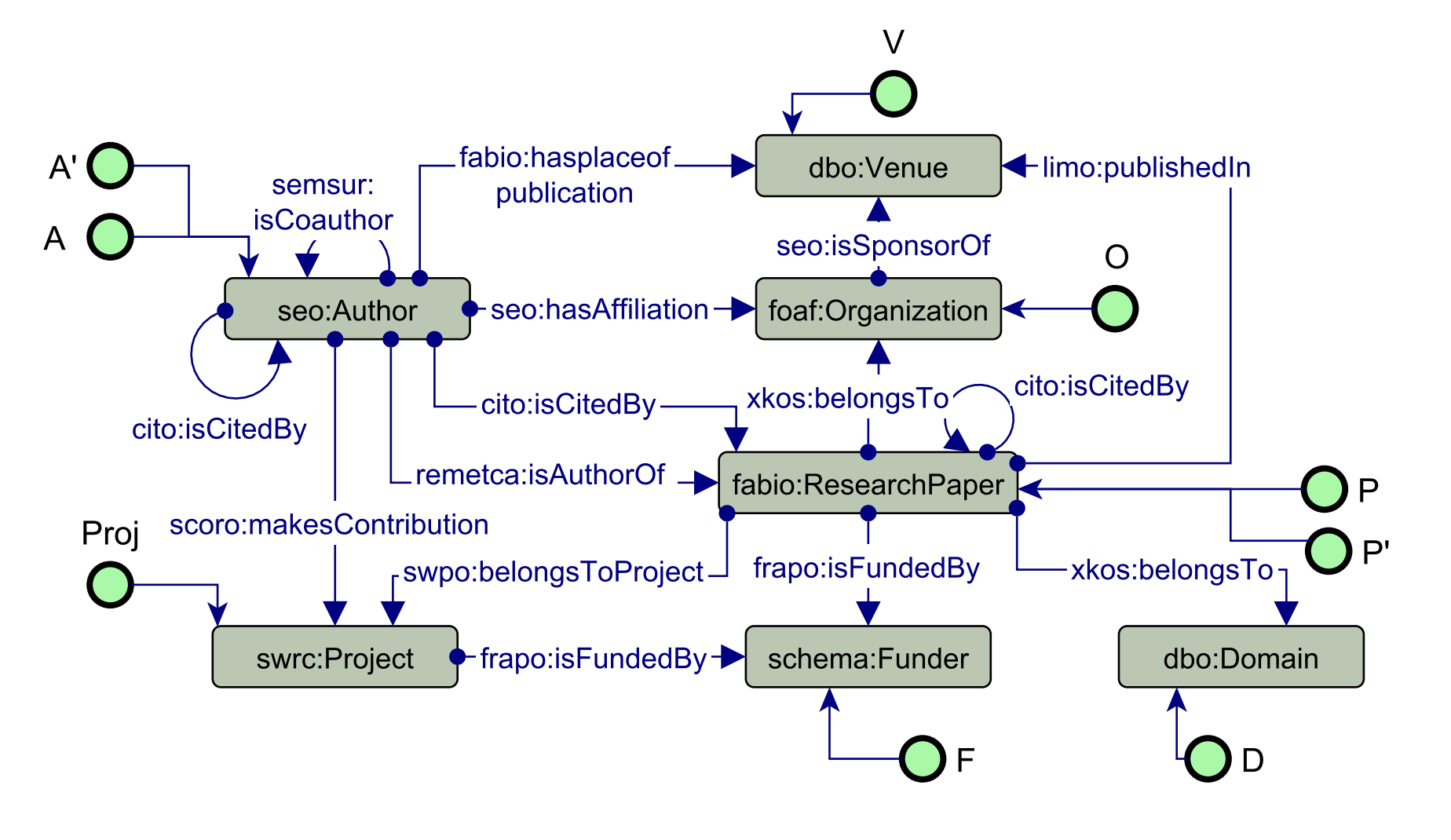
We used PyKEEN to train a scholarly recommendations system to suggest papers to read, grants to apply to, and collaborations to make.
-
Pathway Crosstalk Predictions
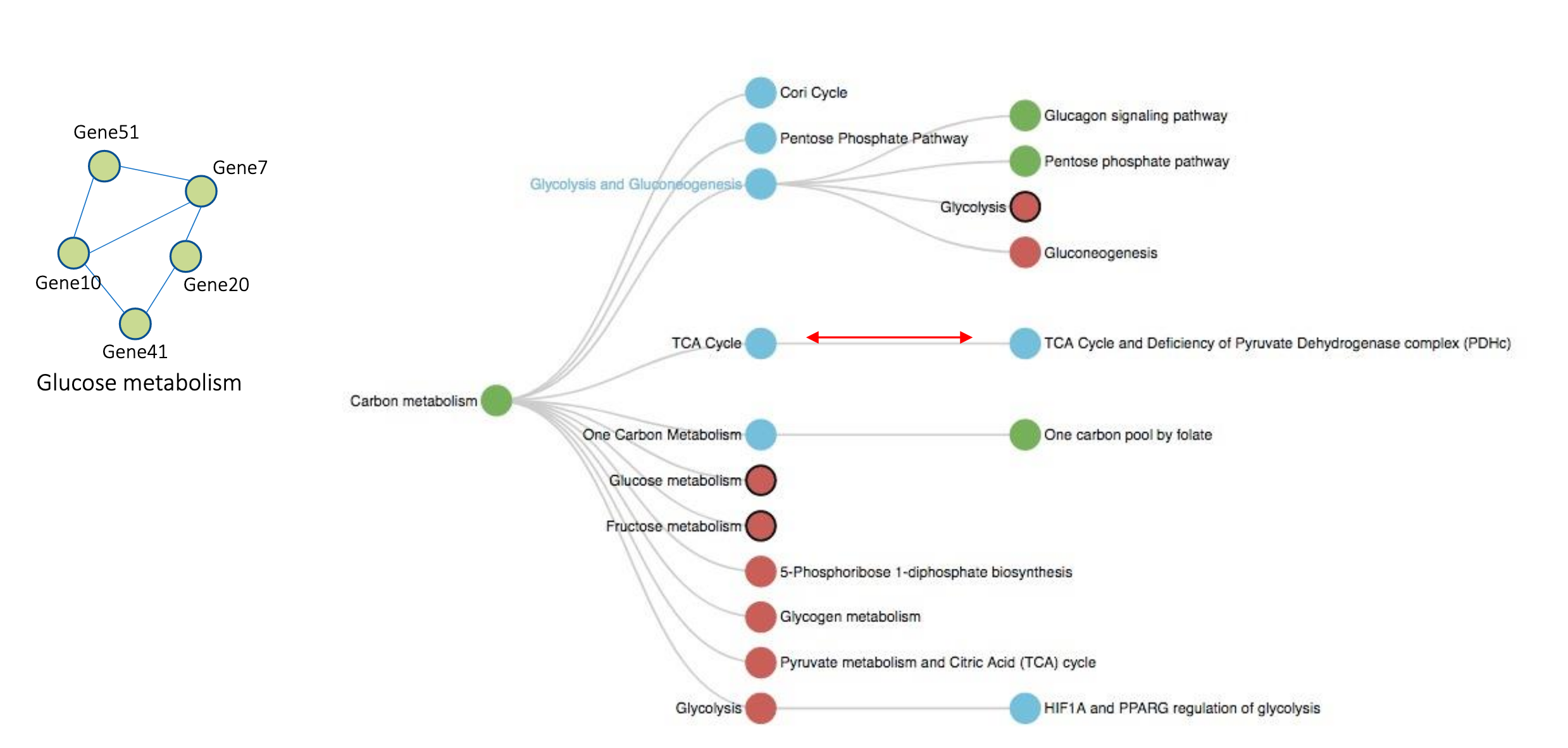
We used PyKEEN to train a pathway crosstalk analysis platform that identifies which biological pathways are connected, giving further insight into normal human pathophysiology and potentially leading to novel hypotheses for understanding the aetiology of complex disease leading to novel drug discovery.
subscribe via RSS